Just as the researchers turned to order one of the designed primer pairs, a well-known scientist from the neighboring laboratory entered the room. After listening to their research plan he said: “Your plan is definitely good and will the answers you are looking for, but there is a faster and cheaper alternative approach to determine people’s genotype, meaning, to distinguish a carrier from a healthy subject. Simply design the primers so that the PCR products from a healthy subject will be significantly different in size compared to the PCR products from a patient. You will then be able to run and separate the products on a gel through electric field (electrophoresis) and… that’s it! You will not need to determine the sequence of the PCR products, which is an expensive and time consuming step… simple and easy. Good luck friends, I must go back to my lab.” The researchers were enthusiastic about the simplicity of his idea and decided to examine its feasibility. They discussed it for a while.
15. Considering the characteristics of the mutation, will PCR amplification products of a normal CFTR and the F508del mutant alleles, using the primers previously designed, located at both ends of the mutation site, differ in length enough to differentiate between them on a gel?
- No, because the amplification product of the mutant allele is shorter in only 3 nucleotides (CTT) from the amplification product of the normal allele, a non-significant difference that will not be noted on a gel.
- No, because the amplification product of the mutant allele is shorter in only one nucleotide (F) from the amplification product of the normal allele, a non-significant difference that will not be noted on a gel.
- No, because there is absolutely no difference between the amplification products of the mutant and normal alleles.
- Yes, because the protein sequence encoded by the mutant allele is different from the normal protein, and therefore a difference between the amplification products will be visible.
The answer is: A. No, because the amplification product of the mutant allele is shorter in only 3 nucleotides (CTT) from the amplification product of the normal allele, a non-significant difference that will not be noted on a gel.
Since the mutant allele harbors a deletion mutation of only 3 nucleotides, the amplification products of the previously designed primers (located on both ends of the mutation site) are not significantly different in length. Using gel electrophoresis (under standard conditions), one can distinguish between DNA fragments that differ in length of at least tens of nucleotides. Sequences that differ in only single nucleotides will run almost the same distance on the gel and be inseparable.
The disappointed researchers turned to the scientist from the neighboring laboratory and presented their difficulty – while using the primers located on both ends of the mutation site, the amplification product of the normal allele is only 3 nucleotides longer compared to that of the mutant allele. The scientist listened and thought for a while, and then cried: “Friends, this is not a problem at all. Use the difference in the sequences of the normal and mutant alleles to design the primers so that the sequence of one of the primers will include the mutation itself, and consequently only the specific F508del mutant allele that complements both primers perfectly will be amplified, while the normal allele will give no PCR product. Great! Now get to work!” The researchers returned to their room full of hope. In order to differentiate a normal DNA sequence of a healthy subject from a mutant DNA sequence of a CF carrier, they, and you, should plan new primer pairs, of which one primer includes the mutation in its sequence and will thus be specific only to one of the sequences.
16. If for the PCR reaction we use a primer pair, of which one primer includes the mutation site, a DNA template from a healthy subject, and the other necessary PCR components – should we expect an amplification product?
- Yes.
- No.
The answer is: B. No, because the primer that complements the unique sequence of the mutant allele (at the mutation site) will not anneal to the DNA template from the healthy subject.
17. In order to get a PCR product, what components should be added to the tube (aside from the Taq polymerase enzyme, buffer and nucleotides)?
- A pair of primers, of which one includes the mutation site and is complementary to the sequence of a mutant allele, and DNA from a healthy subject
- A pair of primers, of which one includes the mutation site and is complementary to the sequence of a normal allele, and DNA from a CF carrier.
- A pair of primers, of which one includes the mutation site and is complementary to the sequence of a mutant allele, and DNA from a CF carrier.
- It makes no difference which primers or DNA we use – the PCR always results in an amplification product.
The answer is: C. For PCR amplification of a target sequence, both primers must complement the template sequence. Only if each of the primers can anneal to a template strand, a significant PCR amplification of the DNA sequence segment that lies between the primers occurs.
18. Will the strategy proposed by the neighbor scientist, meaning, using a primer that includes the mutation site, allow the researchers to distinguish between normal DNA (from a healthy subject) and mutant DNA (from a CF carrier) using gel electrophoresis without the need to determine the DNA sequence?
- Yes.
- No.
The answer is: A. Yes.
Teachers: At this point you can have a discussion in class, reviewing together the different strategies proposed by the scientist from the neighboring laboratory, and see if the students come up with other approaches. It is advised to be open to the different ideas, but lead them to realize that the “mutation-dependent amplification” is the most appropriate strategy. You can refer them to additional reading (pages 129-131 in the textbook Genetic Engineering – from Principles and Methods to Research and Applications). In this method, one of the primers is designed to complement the mutation site, and is specific to the sequence of the mutant allele, and thus an amplification is only possible if the DNA template is from a subject that carries the specific mutant allele (in this case, F508del), while from a normal allele, no amplification product is obtained. Alternatively, we could design a primer to the mutation site that matches to the sequence of the normal allele (meaning, that includes the CTT nucleotides that are missing from the mutant allele), and a product will then be obtained only from the normal allele but not from the mutant allele.
This approach is called “mutation-dependent amplification” and it utilizes “Allele-specific oligonucleotide” (ASO) (Figure 3). It uses a quick and easy method, developed to previously characterized point mutations. The method allows DNA amplification only if the sequence harbors the mutation. A special primer is designed, that has at its 3’ end the unique sequence of the mutant allele, that is different from the corresponding sequence of the normal allele. The second primer anneals to both, mutant and normal alleles, but in order to get an amplification product, both primers must bind the template! Thus, an amplification product is obtained only from a mutant DNA.
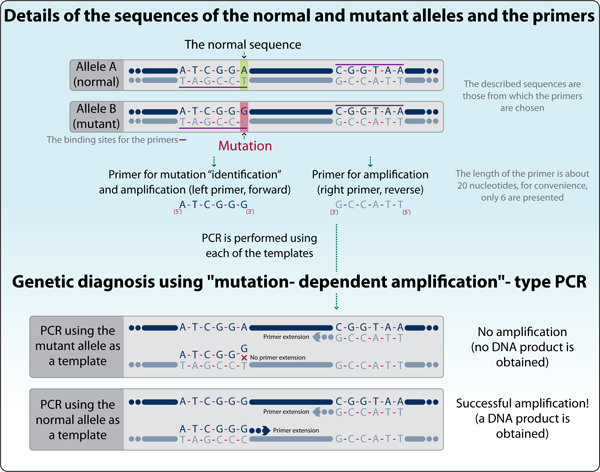
Figure 3: Detection of a specific gene mutation using “mutation- dependent amplification”-type PCR.
For this purpose, we should design a pair of primers of which one primer includes the sequence of the mutation site at, or near, its 3’ end. Here is the sequence of the CFTR F508del mutant allele. It is the same DNA segment we used before to design the primers, but the nucleotides CTT in positions 201-203 of the normal allele are now deleted. Copy and paste the sequence to the appropriate window. In order to dictate the sequence of the left primer so that it includes the mutation site, type in the desired sequence for the primer in the window “Pick left primer or use left primer below” at the bottom of the screen (Screen 8).
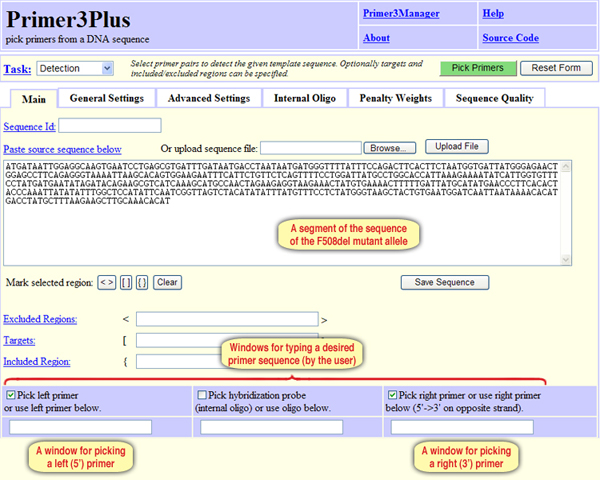
Screen 8: The Primer3Plus tool allows users to determine the sequence of one of the primers or both
19. Assuming the right primer (Reverse) is located at the 3’ end of the segment destined for amplification, right towards its end, at which positions should the left primer (Forward) be located to allow mutation-dependent amplification from the F508del allele?
- The left primer should be located at positions 1-24 in order to get the longest amplification product possible.
- The left primer should be located at positions 230-254 in order to get an amplification product of about 200 nucleotides.
- The left primer should be located in positions 180-202 so it harbors the deletion mutation site.
- The location of the primer is meaningless; for any location we get a significant difference in the size of the amplification products between the normal and mutant alleles.
The answer is: C. Usually, in a mutation-dependent amplification, the 3’ end of the left primer harbors the sequence unique to the mutant allele. Given that in our case the mutation of the F508del allele lies in position 201, the end of the primer must include the nucleotides from this position and forward. The other suggested primers, either in positions 1-24 or 230-250, are complementary to the sequence that is similar between the normal and mutant allele, and therefore amplification products for both the healthy subject and the carrier will be obtained. In the first case, the amplification products will be different in length in 3 nucleotides, whereas in the second – identical in length, and in any case there will be no difference significant enough to be identified using gel electrophoresis. Mutation-dependent amplification, in which a product is obtained only from DNA that harbors the mutation, can work only when the primer is located at the mutation site itself.
When the users dictate the sequence of one of the primers, Primer3Plus checks that it meets to the settings of primer length, annealing temperature etc. (as detailed in the tab “General Settings”). If the primer entered by the user does not meet the settings, an error notification pops up. For example, entering a primer that is too short will be notified by: “Primer too short;” a notification “Tm too low” will pop up if the annealing temperature of the primer is too low. If the primer entered by the user does not have a complementary sequence in the template sequence, the tool will notify: “Primer not in sequence.” The notification “High end self complementarity” denotes that the 5’ and 3’ ends of the primer sequence are complementary and might stick to each other instead of to the template DNA.
20. Considering that the primer length is at least 18 nucleotides, and that it includes the mutation site at its 3’ end, presented are 4 optional primers. Copy each of them and paste to the window “Pick left primer or use left primer below,” click “Pick Primers” and find out for which option a primer pair can be designed without receiving an error notification (the latter will pop up in red-orange at the top of the screen: Left primer is unacceptable)
- GCACCATTAAAGAAAATATCATTG
- GCACCATTAAAGAAAATATCATTC
- GCACCATTAAAGAAAATATCATTGG
- CACCATTAAAGAAAATATCATTGGTG
The answer is: C. We have seen that a change of even one nucleotide in the primer sequence is enough to make the difference between a poorly recommended primer and an optimal one. We should refrain from using primers with an annealing temperature that is too low (“Tm too low”), like primer A; from using primers that do not match the template sequence (“Not in sequence”), like primer B; or from primers which 3’ and 5’ ends have complementary sequences that can stick to each other (“High end self complementarity”), like primer D.
Choose the appropriate primer from the previous question, for which no error notification was received. Copy it again to the window “Pick left primer or use left primer below,” and click “Pick Primers” to assess its qualities (to see if the primer meets specific criteria) and to design pairing primers. All the five primer pairs that were designed share the same left primer sequence (the one we enter), while the sequences of the right primers are different (Screen 9).
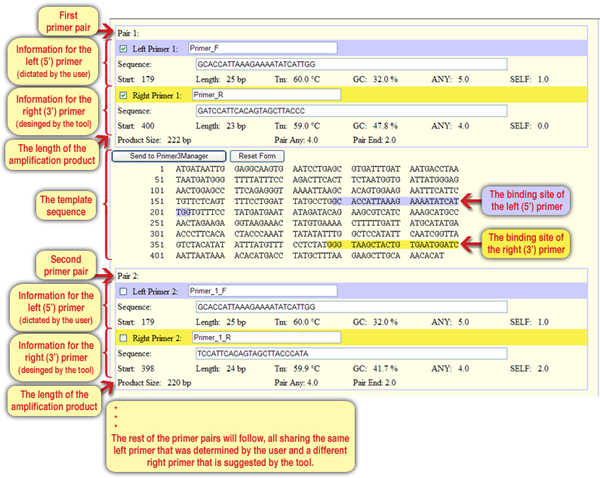
Screen 9: The results screen – designing diverse primers (right primer) to match the primer dictated by the user (the left primer)
21. What can we say about the primer pairs the tool designed?
- They all answer the settings of the tool in terms of primer’s length, annealing temperature, GC type nucleotides percentage etc.
- The right primer is located around positions 370-400 in the sequence.
- An amplification product of about 200 nucleotides is expected from PCR using each of the primer pairs and the F508 mutant allele as a template, but not the normal CFTR allele.
- All the answers are correct.
The answer is: D. All the answers are correct.
Teachers: In mutation-dependent amplification it is not necessarily the left primer (also called the 5’ primer, or forward) that has to include the mutation site. The right primer (also called the 3’ primer, or reverse) might alternatively include the mutation site. Primer3Plus allows the user to dictate the sequence of either primers. . In the latter case, the right primer sequence should be entered to the window “Pick right primer or use right primer below,” and this sequence should be complementary to the sense-strand sequence, since it is a right primer. Alternatively, we can take an approach of amplification that depends on the normal sequence, using the sequence of the normal allele to design the primer. We can dictate the sequence of either the right or left primer to include the mutation site at its 3’ end, but complementary to the sequence of the normal allele. In this case, we will obtain a PCR product from the normal allele but not from a mutant allele (contrary to the expected results of the example described above). It should be noted that mutation-dependent amplification yields a product only from amplification of a specific mutant allele. If no product is obtained, we can assume to have two normal alleles. Other, less likely, interpretations, can be that the PCR did not work at all, or that the site in the template to which the primers should match harbors another mutation. For that reason, we should always use a DNA sample of a known carrier as a positive control to make sure the reaction worked. In addition, along with the PCR reaction that uses mutation-specific primers, a reaction that uses normal allele- specific primers is performed to make sure it is a normal allele.
The researchers chose the first pair of primers the tool offered for the chosen left primer sequence. The left primer sequence starts at position 179 and is 25 nucleotides long, and the right primer starts at position 400 and is 23 nucleotides long. The expected product for a mutant allele template is 222 nucleotides long, while from a normal allele no product should be obtained.