The importance of calcium ions in stabilizing the protein structure
In the cyclic loop structure characterizing subunit A, there is a sequence motif of 19 amino acids which is xGTGNDxuxuGGxuxGxux with each letter representing a certain amino acid or a group of amino acids:
G = Glycine – hydrophobic amino acid;
T = Threonine – hydrophilic amino acid;
N = Asparagine – hydrophilic amino acid;
D = Aspartate – hydrophilic amino acid;
u = hydrophobic amino acid (Valine, Leucine or Isoleucine);
x = some kind of amino acid, usually hydrophilic.
The motif is structured so that the first 10 amino acids (xGTGNDxuxu) are situated opposite the final 9 amino acids (GGxuxGxux).
14. Which part of the sequence motif can be expected to bind to water molecules in the ice crystals?
Choose one answer:
- The sequence xGTGNDxuxu, because it has more hydrophobic amino acids.
- The sequence GGxuxGxux, because it has more hydrophilic amino acids.
- The sequence xGTGNDxuxu, because it has more hydrophilic amino acids.
- None of the sequences, because both do not have any hydrophilic amino acids at all.
The correct answer is: C. In the sequence xGTGNDxuxu there are mainly hydrophilic amino acids T, N, D, x, which constitute 60% of the amino acids in the sequence, and they can bind to water molecules in the ice crystals. On the other hand, the amount of hydrophilic amino acids in the sequence GGxuxGxux is only 44%.
We will now focus on one unit in the ring-like structure made of two loops and two segments of pleated beta sheets. An example of this unit is the unit comprised of the amino acids at positions 77-95. In the first stage we will present only those amino acids by hiding the rest of the amino acids. Perform the following:
- To select the amino acids at positions 1-76 in subunit A, open the console by right clicking the mouse and picking console in the additional options menu. In the console window, right after the $ sign, type Select 1-76:A and press Enter (392 atoms have been selected).
- To hide the part of the structure formed by these amino acids, right click the mouse in the workspace and pick Style -> Structure -> Off. Notice that it is indeed difficult to see the change, but now the amino acids at positions 1-76 are not displayed in the model!
- In a similar method, select the amino acids at positions 96-495 in subunit A using the command Select 96-495:A in the console window and hide them as well (pick Style -> Structure-> Off in the additional options menu).
- Now only the relevant segment in subunit A is displayed (screen 13).
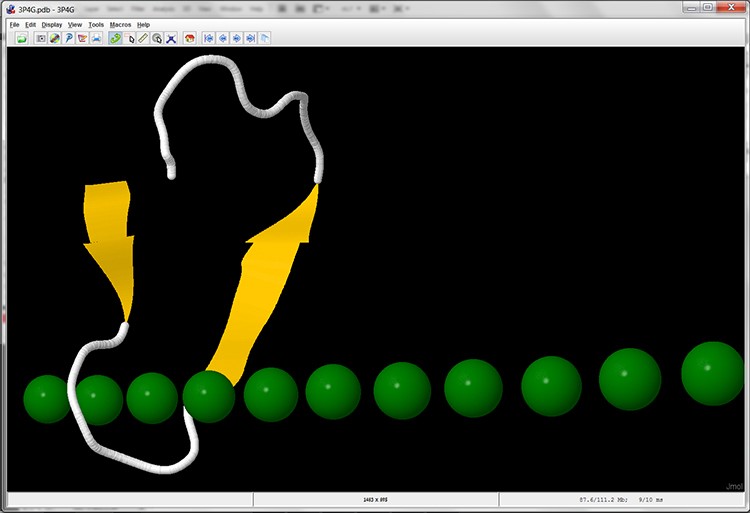
Screen 13: Cartoon display of one unit with a ring-like structure in subunit A of the MpAFP protein. In this display the calcium ions are marked in green.
In the next stage we will try to mark the amino acids in the structure model, which according to their biochemical attributes (meaning, hydrophilic), are believed to be involved in binding to the calcium ions and in this way contribute to antifreeze activity.