First steps
The MpAFP protein is an especially large protein. In an early study it was found that one area of the protein, which is known as area 4(IV) and the length of each of its units is 322 amino acids, provides the protein with its ability to bind to the nuclei of the ice crystals. Therefore many scientists focused only on this part of the protein and experimentally determined its structure.
The information regarding the protein structure is in file 3p4g.pdb. The file extension “.pdb” indicates that this is a molecule structure file.
In order to see the file information in text, it can be opened in “.pdf” format. Examine the open file and scroll down until the lines beginning with – ATOM 1 (an example is seen in screen 1). The letters and numbers displayed represent the type of the atom, its affiliation to a specific amino acid and its location in the three-dimensional space, and in this way a model of the spatial structure of area 4 of the MpAFP protein can be created.
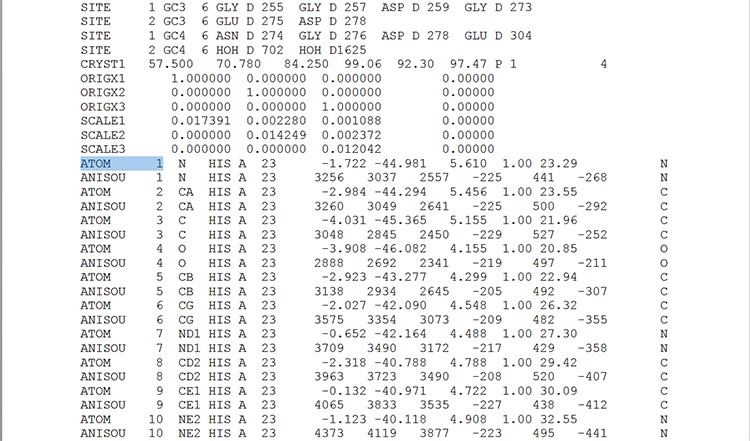
Screen 1: The spatial structure file is composed of numbers indicating the location of each atom in three-dimensional space.
As you can see, the data in the file is not easy to understand and it is difficult to study the protein’s spatial structure using it.In order to view the spatial structure, we must use a special program that can interpret the data in the structure file, and convert the numbers and letters into a spatial shape.
- The purpose of the Jmol tool is:
- To determine the three-dimensional structure of the protein.
- To effectively search for protein structure files in the database.
- To display a three-dimensional model of the protein structure.
- To compare between protein sequences.
The correct answer is: C. The Jmol tool is used to display three-dimensional models of molecule structures such as: proteins, nucleic acids, salt crystals, chemical compounds or any molecule that has a structure file in the database of protein structures.
The Jmol tool receives as input molecule structure files.
Return to the Jmol folder and find the file titled Jmol.jar. This file has a mug icon. Click this file and the Jmol workspace will open.
In the Jmol workspace you can find the main menu bar, while right clicking the mouse will open the additional options menu (screen 2).
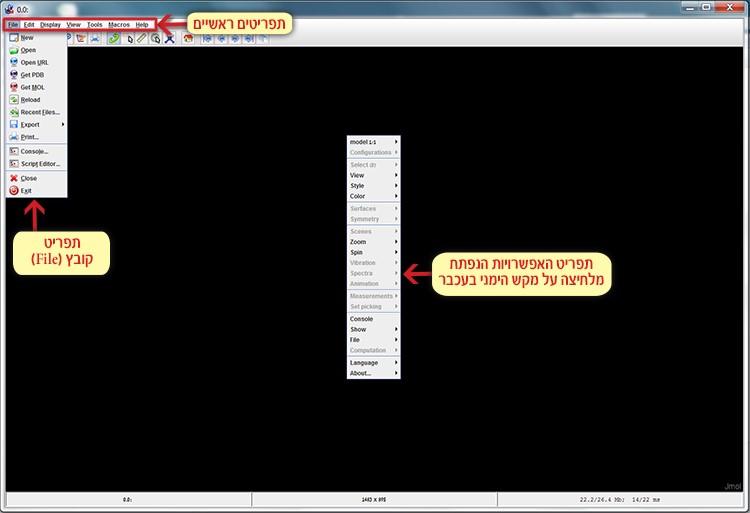
Screen 2: The menu bar (top left) and the additional options menu (on the right) opened after right clicking the mouse.
Now we will open the structure file 3p4g.pdb of area 4 in the MpAFP protein using the Jmol tool:
File ➔ Get PDB ➔ 3p4g.pdbChoose File in the main menu, click Get PDB and in the dialog box that appears, enter the code of the requested file 3p4g (screen 3). This way the file can be opened directly from the database online.
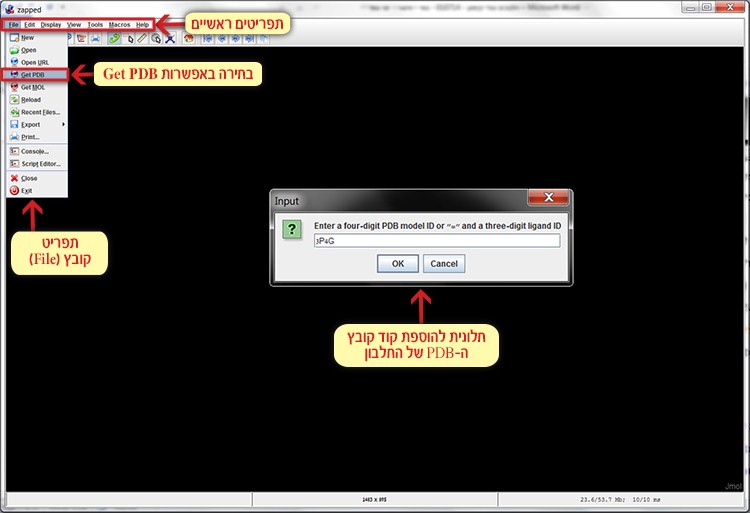
Screen 3: Opening the structure file 3p4g.pdf of area 4 in the MpAFP protein.