In the upper part of the results page, general information about the query sequence is displayed: its name, length and detailed sequence (Screen 2).
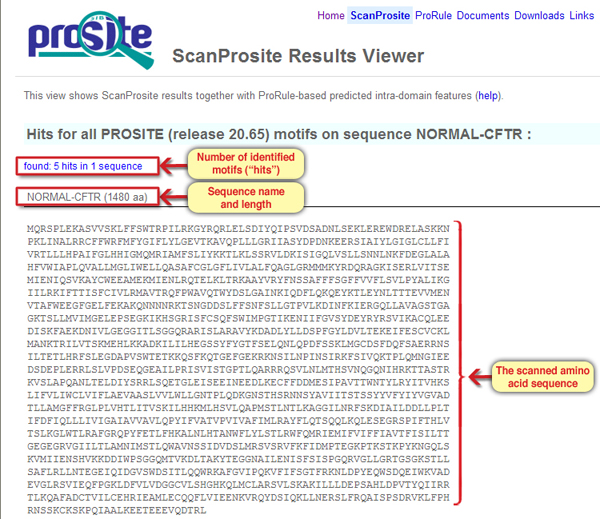
Screen 2: The results page – general information about the query sequence
In the main part of the results page, structural or functional motifs found in the query sequence are represented both in a text form and a graphic form (Screen 3). Each motif the tool found is mentioned by its identification symbol, name, position within the query sequence and a score that measures the sequence similarity between the sequences as appeared in the query protein and the motif as defined in the database. The results page categorizes the results to long motifs (represented by profiles) and short motifs (represented by patterns). Unless mentioned otherwise, we will focus on the long motifs for the purpose of this module.
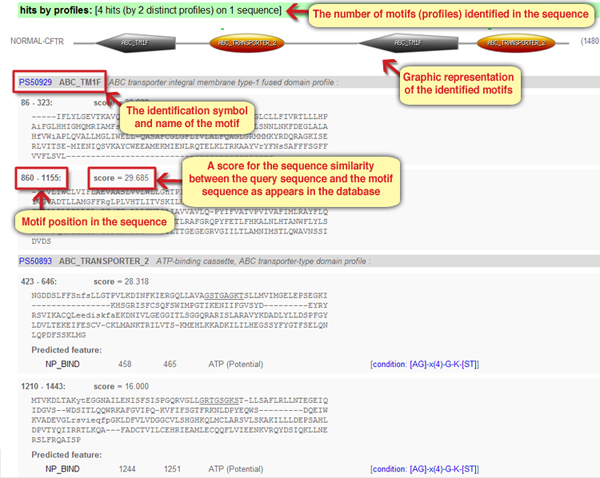
Screen 3: The results page – graphic and text form representation of the long motifs (profiles) that were identified in the protein sequence
Review the results page and answer the following question:
1.How many motifs (profiles) were found in the CFTR protein sequence?
- Two types of motifs (ABC_TRANSPORTER_2 and ABC_TM1F), each appeared twice.
- Four types of motifs (as described, each with its own position, score and unique sequence).
- One motif (ABC), appeared four times.
- Two types of motifs (ABC_TRANSPORTER_2 and ABC_TM1F), each appeared four times.
The answer is: A. Two motifs were identified in the CFTR protein sequence, ABC_TRANSPORTER_2 and ABC_TM1F, each identified twice along the sequence.
2. In which position within the CFTR protein sequence does the motif ABC_TM1F appear?
- It appeared once, in positions 86-323.
- It appeared twice, in positions 86-323 and in positions 860-1155.
- It appears four times, in positions 86-323, 860-1155, 423-646 and 1210-1443.
- Such a motif does not appear in the protein at all.
The answer is: B. The motif appears twice, in positions 86-232 and 860-1155.
In addition, Prosite scores the sequence similarity between the sequence of the motif identified in the query sequence and the motif sequence as deposited in the database. A higher score indicates higher sequence similarity, constituting a motif occurrence.
3. The motif ABC_TM1F was identified twice along the CFTR protein sequence. What can be inferred from the similarity between the sequences in the protein and the motif sequence?
- The sequence in positions 86-323 resembles the ABC_TM1 motif sequence a little more than the sequence in positions 860-1155.
- The sequence in positions 86-323 resembles the ABC_TM1 motif sequence a little less than the sequence in positions 860-1155.
- The sequences in positions 86-323 and 860-1155 resemble the ABC-TM1 motif sequence equally.
- It cannot be determined which of the sequences resembles the motif better.
The answer is: A. The sequence in positions 86-323 resembles the ABC_TM1 motif sequence a little more than the sequence in positions 860-1155.
The names of the motifs appear in short but they usually shed some more light on structure and function. Here, both motifs’ names begin with ABC, indicating that they both belong to the same family.
4. Based on the info line that follows the ABC_TM1F motif name, in which cellular compartment or organelle would you expect to find the motif?
- In the cytoplasm.
- In the nucleus.
- In the mitochondria.
- In the cell membrane.
The answer is: D. It is indicated that the protein is anchored to the cell membrane (“integral membrane”), functioning as a channel for the transport of molecules (“transporter”).
Clicking on the motif identification symbol, a detailed datasheet pops up. In order to learn more about the motif ABC_TRANSPORTER2 lets click on the link PS50893 that appears just left to the motif name. In the datasheet (Screen 4), we can find a description of protein families that share the motif, data on the mechanism of action and the three dimensional structure of the motif (if known) as well as names of other proteins that harbor the motif (Description Section). The main information is usually provided in the opening paragraph, followed by some technical information about the motif and its characteristics (Technical Section) and list of relevant literature (References section).

Screen 4: The opening paragraphs of ABC_TRANSPORTER motif datasheet
Considering the names of the two motifs and the marked inforamtion (underlined in red):
5. Which of the motifs, ABC_TM1F or ABC_TRANSPORTER_2, can bind ATP? What for?
- ABC_TM1F motif binds ATP. ATP breakdown (hydrolysis) releases energy required for various processes.
- ABC_TM1F motif binds ATP that is transported into the cell.
- ABC_TRANSPORTER_2 binds ATP that is then transported through ABC_TM1F out of the cell.
- ABC_TRANSPORTER_2 motif binds ATP. ATP breakdown (hydrolysis) releases energy required for various processes.
The answer is: D. ABC_TRANSPORTER_2 motif binds ATP. ATP breakdown (hydrolysis) releases energy required for various processes.
Lets return to the main results page. Aside from the details on the motif, its position in the query sequence and the score for the sequence similarity, the corresponding sequence in the query protein is also given. Lets look at the sequences in the CFTR protein that were found to resemble the motif ABC_TRANSPORTER_2 (Screen 5). In each of the two corresponding sequences several amino acids are underlined. These are amino acids within the motif that convey an essential activity. Their exact position and function are described under “Predicted Feature”.
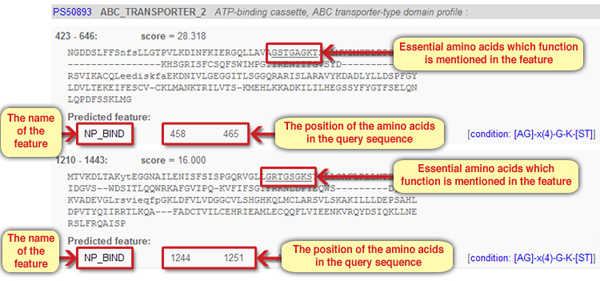
Screen 5: Features in the ABC_TRANSPORTER motif sequences in the protein CFTR
6. In light of the information on the function of the ABC_TRANSPORTER2 motif, and considering the name of the feature, NP_BIND (short for Nucleotide-phosphate binding region), what is the role of the essential amino acids in the motif?
- Transport of metal ions.
- Binding of an ATP molecule.
- Binding of a metal ion, essential for the structure of the motif.
- Stabilization of the structure of the motif.
The answer is: B. The underlined essential amino acids are involved in the binding of an ATP molecule. As we learned, the ABC_TRANSPORTER_2 motif binds ATP, and the feature NP_BIND describes the region or the amino acids within the motif that are directly involved in binding ATP, which is, as we know, a nucleotide.
Comparing the query sequence to the motifs database revealed that the CFTR protein also harbors a short motif (ABC_TRANSPORTER1), represented by a pattern.
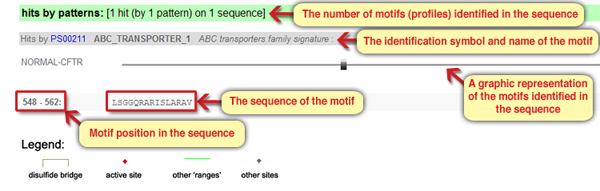
Screen 6: Results page – text form and graphic form representations of the short motifs (patterns) identified in the protein sequence
7. What can we learn from the name of this motif and its positions in relation to the long motifs we learned about before?
- The motif has a structural significance for anchoring the protein in the cell membrane and is a part of the ABC_TM1F motif.
- According to its positions within the sequence, it is a part of the first ABC_TRANSPORTER2 motif, and probably has a functional significance in the binding or dissociation of ATP.
- Short motifs (represented by patterns) and long motifs (represented by profiles) are not related.
- According to its positions within the sequence, it is a part of the second ABC_TRANSPORTER2 motif, and probably has a functional significance in binding or dissociating ATP.
The answer is: B. According to the positions within the sequence it is located as part of the first ABC_TRANSPORTER2 motif, and is likely to be significant functionally, in the binding or dissociation of ATP.
Teachers: This example demonstrates how short motifs are also significant for protein function, and that they can be a part of a longer sequence that is a motif of structural significance. Here we observe a very interesting case, that a comparatively large structural motif, of more than 200 amino acids (ABC_TRANSPORTER2) comprises short motifs of a few single amino acids each, essential for ATP binding (the feature NP_BIND) and its hydrolysis (the pattern ABC_TRANSPORTER1).
Leave the tab of CFTR protein Prosite results page open for a later stage of this task.