The secondary structure display is only a schematic illustration that displays, in the protein’s 3-D structure, the basic structural units: alpha helices, beta sheets and turns and loops; and presents the location of these units in reference to each other. However, we must remember that the protein structure is not visible in reality as helices and sheets. This is a model that displays the protein structure in different ways. If we were able to observe the protein in its natural environment, we would usually see a molecule very tightly folded into a compact round shape.
We will now display the protein in a way that teaches us about its spatial structure and the volume of the atoms that compose it. Right click the mouse in the workspace. In the additional options menu, pick Style ➔ Scheme ➔ CPK Spacefill.
This is the spatial atomic display of the protein. Each atom is displayed as a sphere, while the volume of each sphere and the distance between them represents the volume of each atom and the distance between the atoms. That is why this display is also called atomic volume display. It is also common to refer to this display as Van der Waals interactions display, because the size of the electron cloud (the volume of the sphere) and the surface area are among the main factors (along with polarity) influencing the intensity of the Van der Waals interactions existing between the atoms and between the molecules.
The Select command
Some of the red spheres represent the oxygen atoms in the water molecules, which surround the protein. We will omit them from the model. In order to achieve this we will use the Select command, which enables us to select part of the model. This command requires two stages as shown in Figure 7: using the Select command we will select the component or part of the molecule on which we will perform the action, in this case the water molecule; and on the next stage we will omit the water molecules from the model. Return to the workspace and perform the following actions:
- Right click the mouse in the workspace. In the additional options menu
- Select ➔Hetro ➔ All Water.
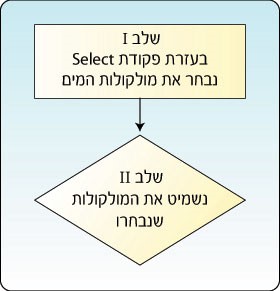
Figure 7: Performing an action on part of the protein requires two stages. In the first stage select the desired part of the protein and in the second stage perform the desired action
- Did the protein structure display change after selecting this command?
- Yes
- No
The correct answer is: b. Note that after the Select command was performed there was no change in the model display, since we chose a certain part of the protein – in this case the water molecules surrounding the protein – but have not performed an action that would change the protein display.
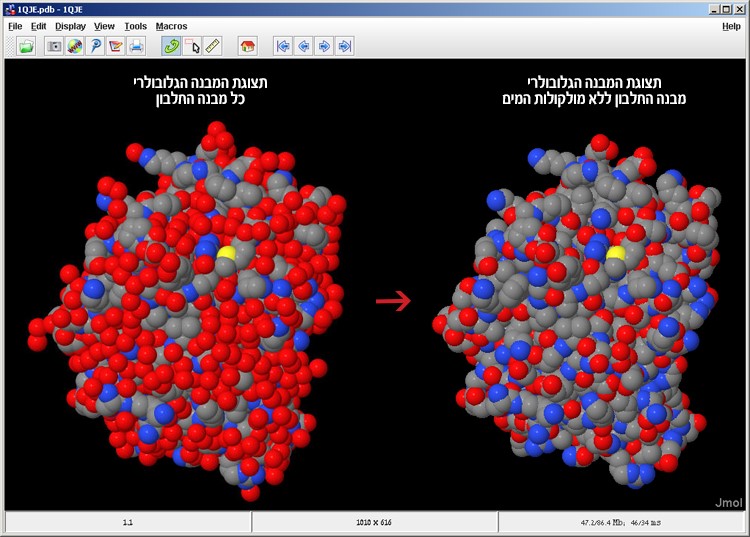
Figure 8: The IPNS protein model in the CPK Spacefill display before (left) and after (right) omitting the water molecules from the model
- Right click the mouse and select Style ➔ Atoms ➔ Off
The water molecules have been removed from the model and now the protein’s atomic volume display is visible to us, without the water molecules surrounding it (Figure 8).
Perform the command required in the feedback for question 5
- Are alpha helices or beta sheets visible in the CPK spacefill model display?
- Yes
- No
The correct answer is: b. Secondary structures cannot be identified in any way in the CPK spacefill display.
- Most of the water soluble proteins have a globe-like structure. This is why they are called globular proteins. Water solution containing globular proteins will appear as a clear solution. Discuss this amongst yourselves and speculate why this is. In your answers address the location in the protein of hydrophobic amino acids, which hate water, and hydrophilic amino acids, which love water.
Globular proteins are water soluble proteins, since the hydrophobic amino acids that hate water are located in the interior of the protein and are not exposed to water; while the hydrophilic (polar) amino acids are usually found on the exterior of the protein and are exposed to the watery environment of the protein. We will color the hydrophilic amino acids in cyan. This action also requires using the Select command since we wish to select only part of the complete model and perform the action on that part only, in this case to color the hydrophilic (polar) amino acids. See Figure 9.
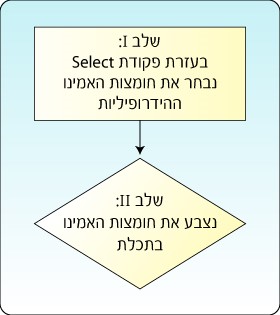
Figure 9: Selecting the hydrophilic amino acids in the model requires two stages
Return to the workspace and perform the following actions:
- Right click the mouse in the workspace.
In the additional options menu, pick Select ➔ Protein ➔ Polar Residues. - Right click the mouse and pick Color ➔ Atoms ➔ Cyan.
We will color the hydrophobic (nonpolar) amino acids white. Return to the workspace and perform the following actions:
- Right click the mouse in the workspace. In the additional options menu, pick
Select ➔ Protein ➔ Nonpolar Residues. - Right click the mouse and pick Color ➔ Atoms ➔ White.
Now the hydrophilic amino acids are cyan and the hydrophobic amino acids are white (screen 6).
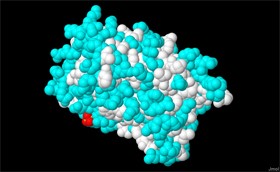
Screen 6: IPNS protein model with the hydrophilic amino acids marked in cyan and the hydrophobic amino acids marked in white
- Observe and rotate the model before you (observe screen 6 as well), and write down which is the most prominent color on the protein exterior, the area exposed to the watery surroundings of the cell, white or cyan? What can be concluded from this? How can this phenomenon be explained?
- Can we identify the location of the active site in the current display of the atomic spatial structure?
- Yes
- No
The correct answer is: b. The atomic spatial structure display, or the protein surface display, mainly enables a thorough examination of the protein surface (the exterior structure), but it does not provide enough information about the protein interior (interior structure). For example, in the atomic spatial structure display, it is very difficult to observe the substrate molecule located in the protein interior, very close to the active site, even though the color of the substrate molecule in the model is not cyan or white. Therefore the CPK spacefill display is not suitable for examining the active site, whose structure is usually like an internal “pocket” in the protein. As stated at the beginning of the task, the active site is composed of three amino acids. In order to identify their location, the scientists must mark them in a way that will distinguish them from the rest of the amino acids in the protein, and in this way the location and structure of the active site can be found.