Teachers: Gene mutations can affect different molecular pathways involved in protein synthesis, processing, transport to the cell membrane, stability and function. The major point is that disease severity is determined by the quantity of normal protein or the functionality of the existing protein. We will discuss different approaches for choosing therapies individualized according to a specific mutation or disease pathogenicity. In the following task, we will work on diagnosing nonsense mutations that lead to early interruption of protein translation. Known mutations of this type are 3849+10kb CàT CFTR or W1282X. There are drugs that allow reading past an early stop codon, the result of the mutation, and into the original stop codon, resulting in formation of a protein with normal sequence and length.
Many mutant CFTR alleles that harbor different mutations are known. Some of the mutations cause a significant change in the sequence of the messenger RNA or the protein sequence. Despite the significant changes in sequence, at times, some of the CFTR activity is maintained. Most carriers of such mutant alleles present a milder phenotype of the disease. Thus, the type of the mutation (deletion, insertion, substitution etc.) and its position within the gene are highly important.
CF is inherited in an autosomal recessive pattern. Thus, in patients both alleles are mutated, and the combination of the two mutant alleles has a great impact on the phenotype. Through the former task, we learned how to use bioinformatics tools to identify the affected motif in the mutant protein. Such findings will also affect the choice of treatment and therapy.
Physicians and researchers are currently trying to develop definitive treatments for CF, to replace today’s supporting therapies aimed only to relieve symptoms. Efforts are devoted to match patient’s care and medications to to the mutations he/she carries. Drugs under development are aimed to address the various types of defects in the process of CFTR protein formation. For example, it was found that administering an aminoglycoside type antibiotic, such as gentamycin, is beneficial to CF patients whose disease is caused by a nonsense mutation (a premature stop codon). This treatment allows the translation machinery to read through the premature stop codon and down to the original one, producing a full-length protein (for more details, click here). Inspired by the clinical improvement of patients treated with gentamycin, non-antibiotics chemichal compounds were developed, such as PTC124, and lead to even better results reduced toxicity and side effects (for more details, click here and also here). In addition, drugs that assist in the folding of a defected CFTR (such as F508del) and its transport to the cell membrane, or drugs designed to improve CFTR function as a channel in the cell membrane, are under study. The way to a genetic mutation- based drug tailoring starts through research that relies on bioinformatics tools that facilitate the study of the qualities of the different gene mutations and their effect on protein structure and function. In the context of mutation analysis, bioinformatics tools can also serve for the diagnosis of carriers, namely, a tool that can diagnose a person who carries a mutant allele. Identification of CFTR mutations is important not only for directing therapies according to the individual’s defect in the mechanism of protein production, but also for family planning consultation for patients and carriers. Couples undergo genetic testing to find out if they are carriers of CFTR gene mutations are then consulted on their chances of giving birth to a child with the disease.
Clinical Case Study
Two tubes of genetic testings were received at the hospital laboratory, each with a sticker and a DNA sample. The genetic councelor attached a letter to the tubes explaining that one tube contains DNA from a healthy subject, with no mutations in the CFTR gene, while the DNA in the second tube is of a CF carrier with the F508del mutation. Unfortunately, the ink in the printer must have run low and the stickers were hard to read. They could read the persons’ names on the tubes but not their status. Now, the researchers (and you) must define which of the two samples is of a carrier, and which from a healthy subject, and to do it quickly and at minimal cost. The researchers decided to sequence the CFTR allele at the F508del mutation region for both DNA samples and compare them to the normal and mutated sequences. However, when they opened the tubes they realized they had too little DNA from each sample, not enough for sequence analysis.
1. Which bioinformatics tool should the researchers use in order to compare the DNA sequence they obtain (after sequencing) to the normal and mutant CFTR alleles?
- Entrez – a search tool based on a text query.
- BLAST – a search tool based on a sequence query.
- Prosite – a tool for the search of structural and functional motifs in a given protein sequence.
- ClustalW – a tool for sequence alignment.
The answer is: D. ClustalW – a tool for sequence alignment.
2. Which method can be used to determine the nucleotide sequence of a DNA molecule?
- Cutting with restriction enzymes.
- PCR (polymerase chain reaction) amplification.
- Sanger’s DNA sequencing method.
- DNA cloning using genomic libraries.
The answer is: C. Sanger’s DNA sequencing method. For more information about this method, watch the attached video that also appears with explanations on Davidson on-line.
3. Which method can be used for the amplification of a defined DNA sequence in-vitro?
- Cutting with restriction enzymes.
- PCR (polymerase chain reaction) amplification.
- Sanger’s DNA sequencing method.
- DNA cloning using genomic libraries.
The answer is: B. PCR amplification. For more information about this method, watch the attached video that also appears with explanations on Davidson on-line.
Thus, the researchers chose to use the polymerase chain reaction (in short, PCR) to amplify a region from the gene sequence that encompasses the mutated area, and then to determine its sequence using Sanger’s DNA sequencing method. By comparing the obtained nucleotide sequences of the DNA samples to that of the normal allele and the F508del mutant allele the researchers hoped they learn which is which.
Teachers: At this point, it can be helpful to go through the stages of the PCR reaction and its applications (for example, you can use the textbook Genetic Engineering – from Principles and Methods to Research and Applications by Dan Michael and Anat Yarden, chapter 6, especially through pages 122-131) and also the principles of sequencing (chapter 5 in the same textbook, especially pages 97-100). The abovementioned videos and animations can also be useful.
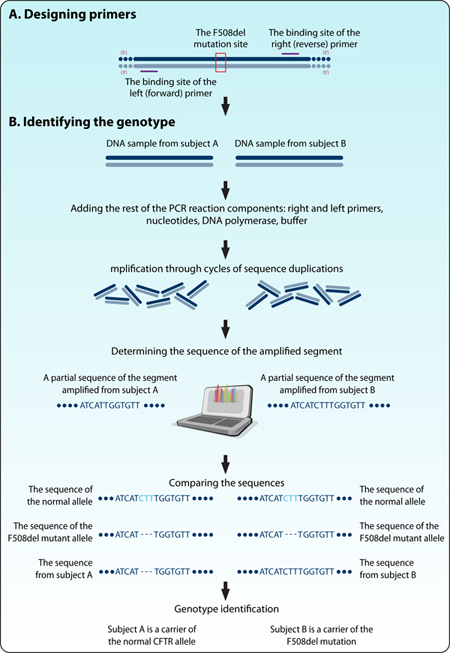
Figure 1: The scientific approach for distinguishing between normal and mutant alleles using PCR, sequencing and sequence alignment.
The goals for this task:
We will begin with designing PCR primers for the diagnosis of F508del CFTR gene mutation carriers using the tool Primer3Plus, and then analyze PCR products and identify the carrier and the healthy subjects.