Lets go back to the main page by clicking “Back.” Oftentimes, an amplification of a segment under specific requirements is necessary. The primer pairs offered by the tool might not match the requirements unless the user defines the requirements precisely.
In the “Main” tab, we can define 3 main limitations (Screen 7), which are:
Excluded regions – Specifies the region in the sequence from which the primers should not be chosen. The number on the left indicates the first position and the number on the right indicates the number of nucleotides from this position and on, meaning, the length of the segment from which primers should not be designed. For example: 151, 200 – the primers will not be chosen from position 151 and for 200 successive nucleotides, meaning, until position 350 in the sequence. Alternatively, we can mark the desired nucleotides on the sequence and click < >.
Targets – Specifices the region in the sequence that will be captured between the primrs. The number on the left indicates the first position and the number on the right indicates the number of nucleotides from this position and on, meaning, the length of the segment that from both sides the primers will be designed. For example: 151,200 – the two primers will be placed flanking the region that starts at position 151 and for 200 successive nuclotides, meaning until position 350. Therefore, the left primer will be positioned before position 151 (somewhere from the beginning of the sequence to 150) and the right primer will be positioned after position 350 (somewhere from 351 and to the end of the sequence). Alternatively, we can mark the desired segment on the sequence and click [ ].
Included Region – Specifices the region in the sequence where primers should be chosen. The number on the left indicates the first position and the number on the right indicates the number of nucleotides from this position and on, meaning, the length of the segment in which the primers will be located. For example: 151,200 – both primers will be positioned in the segment between position 151 and along 200 nucleotides, meaning, between positions 151 and 350. Alternatively, we can mark the desired segment on the sequence and click { }.
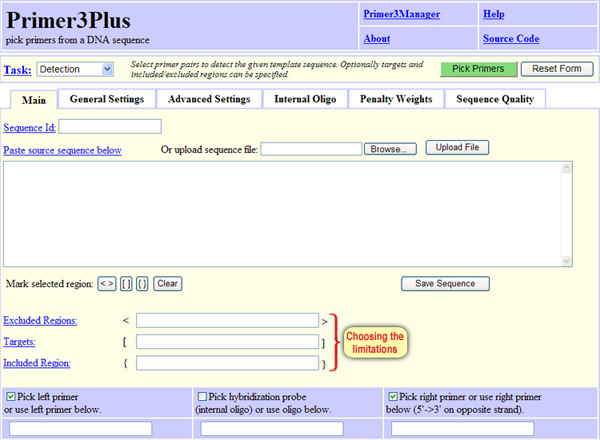
Screen 7: Defining limitation options for primer design
A graphic explanation for limitation definitions can be reviewed in figure 2.
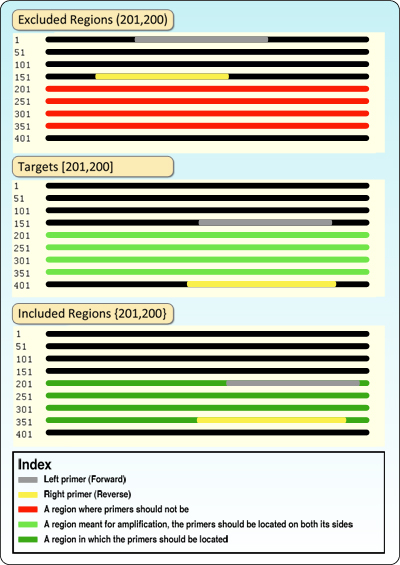
Figure 2: designing primers by using limitation options
13. We are interested in designing primers to amplify the segment that includes positions 201-203 that are missing in the F508del mutant allele. Which is the most appropriate limitation?
- Excluded regions <3,201>.
- Targets [3,201].
- Included Region {201,3}.
- Targets [201,3].
The answer is: D. We want to amplify the segment that includes positions 201-203 so the primers should be located on both sides of it, and therefore, we choose the limitation “Targets.” First (on the left), we indicate the first position in our target, which is 201, then (on the right), the number of nucleotides from this position, which is 3.
Teachers: The limitation in statement A will exclude primers from positions 3-201 and so all primers will be designed from after position 204. The limitation in statement B means that the primers should be located on both sides of the segment from position 3 to 203, but since the primer should consist of at least 18 nucleotides, the tool will not be able to design a left primer as the end of the target segment is too close to the beginning of the sequence. The tool will not be able to design any primers following the limitation in statement C, since a region of three nucleotides is too short to plan from within it two primers (of at least 18 nucleotides each) with an amplification product of 150 nucleotides.
Lets define this limitation condition and click “Pick Primers.” Note that positions 201-203 (nucleotides CTT in a normal allele) are highlighted in green, according to the limitation. Make sure that for all the designed primer pairs the left primer is positioned at the beginning of the sequence and before position 200 (the Start field of the left primer is marked in grey) while the right primer is positioned in the center of the sequence and after position 203 (the Start field of the right primer is marked in yellow).
14. Think and discuss – is this the only limiting condition that can yield appropriate primers for the amplification of the desired segment? Explain! If you come up with other limitations, please write an example.
Teachers: A few appropriate limiting conditions can be written. The simplest one is to choose a “Target” condition of a wider area that includes positions 201-203, for example: Target [201,10] (positions 201-210) or Target [198,8] (positions 198-205) etc. An alternative approach is to use the “Excluded Regions” condition to indicate a relatively large area where primers should not be positioned so not to allow the tool to design primers from the same side of the excluded segment (right or left; given that the minimal length of the amplification product is defined as 100 nucleotide pairs). This will force the tool to design primers from both sides of the excluded region, for example: Excluded Regions <101,250>, one primer will be located between positions 1 and 100 and the other between 351 and 450. Another option is to provide the limitation “Included Region” with a comparatively short region where the primers should be chosen from with the mutation site at its center, which will force the tool to choose primers from both sides of the mutation, for example: Included Region {100,200}, one primer will be located between positions 100-170 and the other between 230 and 299 to allow an amplification product with an optimal length of 150 base pairs.
At this point, the researchers have 5 primer pairs, which can be used in a PCR together with the other essential components (DNA polymerase enzyme, buffer, template DNA and dNTP nucleotides) to amplify the CFTR gene sequence that harbors the site of the F508del mutation in an affected allele. The nucleotide sequence in this segment will be determined using Sanger’s DNA sequencing method. The DNA sequence will be compared to sequences of the mutant and normal CFTR alleles as appear in databases of nucleotide sequences, and its source, whether from a healthy or carrier subject, will be determined.