The PCR allows the cyclic duplication of defined DNA segments in-vitro and significant subsequent amplification. Its high efficiency, sensitivity and simplicity, made the PCR an essential and common tool in different research fields (science, medicine, law).
4. What are the required components for the PCR amplification reaction?
- Template DNA, primer pair and dNTP type nucleotides.
- Template DNA, primer pair, dNTP type nucleotides and DNA polymerase enzyme.
- Template DNA, primer pair, dNTP type nucleotides, DNA polymerase enzyme and the buffer.
- Template DNA, primer pair, dNTP type nucleotides, RNA polymerase enzyme and the buffer.
The answer is: C. The required componnets include: a DNA template that encompasses the region needed to be amplified; a pair of specific primers that bind complementary sequences in the DNA template strands in sites that define the ends of the amplified segment of interest; dNTP nucleotides, the building stones of the DNA; the DNA polymerase that elongates the primers by adding nucleotides according to the sequence in the template DNA strands; and the buffer.
5. A typical amplification cycle of the PCR includes 3 stages. What is their sequence and in what temperatures do they take place?
- Denaturation of strands (at 95°C), annealing of primers (at 50-65°C) and primers elongation and DNA synthesis (at 72°C).
- Annealing of primers (at 50-65°C), denaturation of strands (at 95°C), and primers elongation and DNA synthesis (at 72°C).
- Denaturation of strands (at 72°C), annealing of primers (at 72-95°C) and primers elongation and DNA synthesis (at 95°C).
- Annealing of primers (at 95°C), primers elongation and DNA synthesis (at 72°C) and denaturation of strands (at 65°C).
The answer is: A. First, the DNA strands are separated (denaturation) at a high temperature of 95°C, resulting in single-stranded DNA molecules. Then, the primers bind to complementary sequences (annealing), each primer to a different DNA strand. This happens at moderate temperatures of 50-65°C (the exact temperature is determiined by the sequences and length of the primers) and sets the ends and length of the amplified segment of interest. Finally, the DNA polymerase enzyme uses the single strand DNA as a template to add complimentary dNTPs to the 3’ ends of each primer and generates a section of double-stranded DNA in the region of interest, a stage that mostly takes place at 72°C, the optimal temperature for the activity of the enzyme. After several cycles (usually 20-30), many new copies of the DNA segment of interest are amplified.
As mentioned, from the DNA samples they received, the researchers chose to amplify a specific segment of the CFTR gene that includes the three nucleotides deletion mutation site in the F508del mutant allele. By sequencing the amplified allele segment and aligning it to the normal and mutant CFTR sequences they should be able to determine if the source of the DNA sample is from a healthy subject or a carrier.
In order to design PCR primers to be used for the amplification of the desired sequence, we will use the tool Primer3Plus. Click on the link to open Primer3Plus (Screen 1). In addition to the window where the DNA sequence should be inserted, the tool interface also includes windows to define the purpose of the PCR, primers design clicker and more functions we will get acuaintant with later.
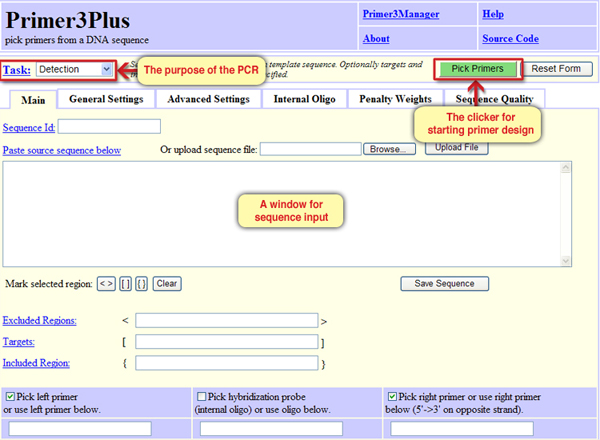
Screen 1: The Primer3Plus interface – a tool for designing primers, to be used for example in PCR method.
We should indicate the purpose of the primers in the “Task” window. Primers can be used for different purposes, such as: cloning, sequencing, sequence detection etc. Different primers can be designed according to specific needs. Lets choose the option “Detection” that means sequence identification (Screen 2), since we wish to identify the source of the amplified DNA – whether it is from a healthy subject or a carrier.
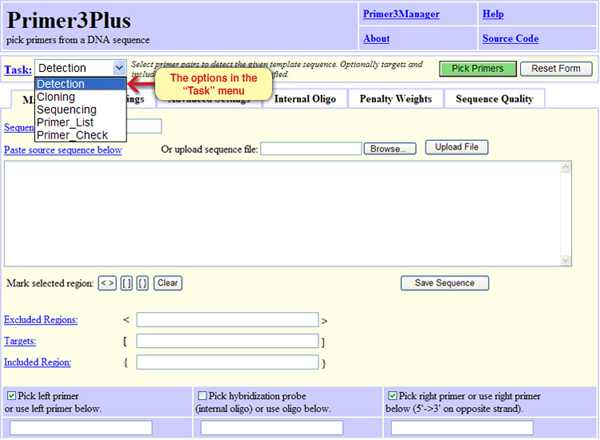
Screen 2: choosing the purpose of the primers.