For teachers: Similar to the way we analyze nucleotides records (see the first task in the module “Mutations Save Lives”), we can analyze records of proteins (see third task in the module “The Arms Race”). Protein records include information about the amino acid sequences that are important for the structure and function of a protein such as an active site of an enzyme, an ion binding site, etc. When studying a protein sequence, we are restricted to proteins registered in the database. Prosite predicts the existence of structural and functional motifs in the studied protein (input sequence) by searching for similarities to sequences formerly experimentally defined and submitted to a motif designated database. Thus, Prosite allows even the analysis of protein sequences that were not registered in a protein sequence database. The input (query) submitted by Prosite users is a sequence of amino acids, independent of its source or whether it is only partial. The tool can be used, for example, to study a sequence of a mutant protein or protein segment isolated in the lab, on which no prior data exists or only partial data can be found in the databases.
How can bioinformatics tools, such as Prosite, assist in studying the structure and function of a protein? A basic assumption in bioinformatics is that similarities between sequences (conserved sequences) usually suggest similarities in structure and function. When we compare the sequence of amino acids of an investigated protein to sequences of proteins which structure and function were studied and characterized in past experiments, and were uploaded to the database, we are able to recognize a protein family the investigated protein belongs to, as well as predict the structure and function of a certain segment of the protein or its whole. Sequence similarities among the studied protein sequence and database sequences suggest protein segments of similar structure and function. Hence, by comparing the protein sequence to a database of protein motifs we are able to identify motifs in the studied protein that endow the protein with a certain structure, function or activity (Figure 2).
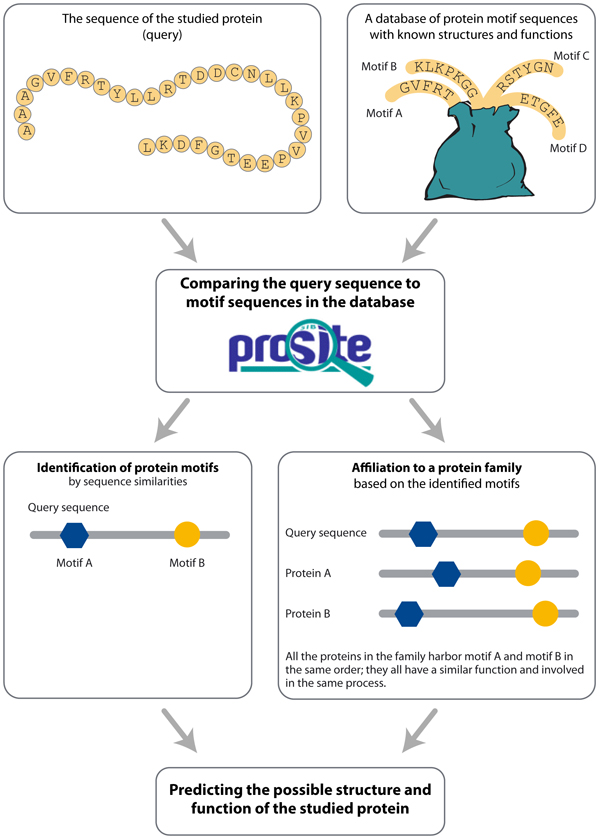
Figure 2: Prosite predicts the motifs that may occur in the studied protein sequence and whether it belongs a protein family, and foresees possible protein structure and function.
The tool Prosite predicts what motifs may occur in the studied protein and whether it is a member of a protein family, based on a sequence similarity search through a designated database of protein motifs and families (Figure 2). We will use Prosite to study which motifs may occur in CFTR protein sequence and their functional importance. We already know that the protein serves as an ion transport channel, but still have some questions open: which motifs allow the formation of the channel in the membrane? Which motifs allow the transfer of ions through the channel? We will analyze whether these motifs are affected by different gene mutations and how it relates to disease phenotype.
Before we continue with the module, it is advised to watch the guided tutorial that explains the basics of using the tool Prosite.