At the beginning of this activity we knew the location of the amino acids comprising the active site of the IPNS enzyme in the amino acid sequence of the protein: Histidine at position 214 (marked H214), Glutamic acid at position 216 (marked D216) and Histidine at position 270 (marked H270). We asked how is it possible that the active site is comprised of three amino acids when one of them, H270, is far from the other two? Is the distance in the amino acid sequence evidence that the active site spreads over a large area of the protein spatial structure?
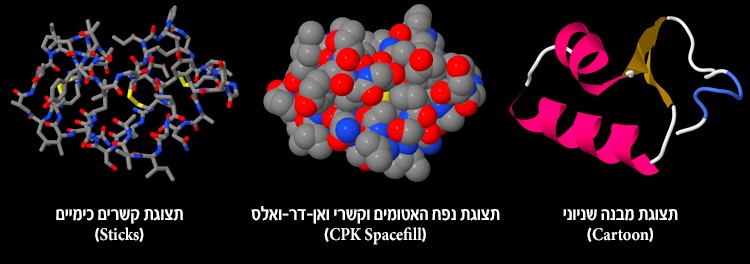
In order to answer these questions and to understand the mechanism of action of the active site, we used the Jmol workspace, which displays spatial structures of molecules. We learned that the protein structure can be displayed in many display options and that each one demonstrates and emphasizes different aspects of the protein. For example, the balls and sticks display presents all of the atoms and all of the chemical bonds between the atoms. Using this display method we can see single amino acids. This way we saw the structure of the active site. However, with this display option it is difficult to distinguish the secondary structures of the protein. That is why we chose the cartoon display, which presents secondary structures. We saw that there are three types of secondary structures in the protein – alpha helix, beta sheet and also turns and loops that connect these structures. The protein can be displayed in other ways as well, which were not mentioned here, but can be examined in the workspace. However, we must remember that these are only different representations of the structure, and this is not necessarily what the protein looks like in reality.
16. Before you are three different display options of the same protein. Which of these options displays the protein structure as it is in reality?
17. We have a different protein with a mutation at position 5, and you suspect this mutation is affecting the activity in the active site. Which display option do you suggest to display the protein structure and how will you check your hypothesis if you know that the active site is located at positions 20, 25 and 37?
At the end of the activity we also examined the active site and its spatial structure and realized that the three amino acids are very close to each other in the spatial structure, even though they are far from each other in the primary structure (the protein sequence) of the protein. We learned that the enzyme catalyzes a chemical reaction in which the chemical bonds in the substrate molecule change, and a product molecule is created with a closed 5-membered ring. Due to the ring forming, the product molecule is shorter than the substrate molecule and changes the angle in the active site. And so, the product molecule detaches from the active site and is released into the watery surroundings. The enzyme’s activity requires electron transfer. This is why, at the active site of the IPNS protein, there is an iron metal ion (Fe+2) that serves as a catalyst.
18. A biology student accidently boiled a test tube with protein solution. The student discovered that after boiling, protein activity ceased. The student assumes that the boiling process affected the protein structure. Which of the following structures were affected by the boiling? The primary, secondary or tertiary structures of the protein? Explain your answer.
19. In many proteins there are disulfide bonds between two sulfur atoms, which help stabilize the protein structure. For example, the Insulin protein has disulfide bonds. The amino acids containing the sulfur atom are cysteine amino acids. Before you is the spatial structure of a protein in the cartoon display, and you wish to determine if it has disulfide bonds. Describe what actions you should perform in order to answer this question (disulfide bonds are displayed in yellow).
In this task, the scientists found that the active site of the protein contains three amino acids that are not next to each other in the protein sequence, and therefore decided to observe the spatial structure of the enzyme. They determined the spatial structure of the IPNS protein, which originates from the fungus Aspergillus Nidulans, and using a tool for displaying protein structure, displayed the structure of the enzyme’s active site, substrate, product molecule and the metal ion,
which serves as a cofactor. Now that the scientists have acquired plenty of information about the IPNS enzyme, they can think of new directions for research in the war against pathogens.
In this task, the scientists wished to characterize the structure of the IPNS enzyme using a tool for displaying molecule structures. The scientists presented the spatial structure of the IPNS enzyme from the fungus Aspergillus Nidulans and focused on the structure and the relative location of the enzyme’s active site, substrate, product molecule and the metal ion used as a cofactor. The use of computerized models, like the tool used by the scientists in this study, may contribute to the arms race between humans and bacteria, which in turn are used in groundbreaking studies searching for new antibiotics and for planning improved antibiotics.