The tool scans the DNA sequence and detects primers that answer a variety of settings and considerations according to the PCR purpose. The main definitions are detailed in the tab “General Settings.” Lets open this tab. Read the default settings as detailed in this tab (Screen 3) and answer the following questions regarding the characteristics of the primers and the expected amplification product.
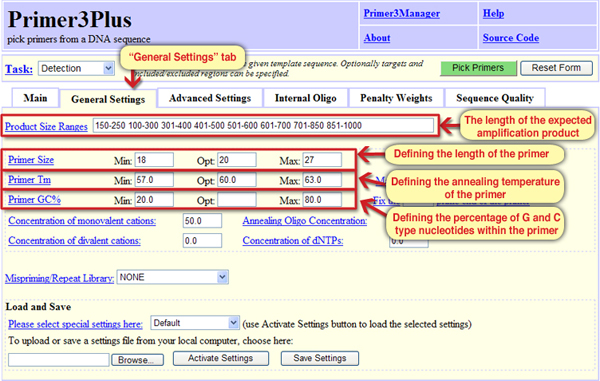
Screen 3: Defining the characteristics of the primers and the predicted amplification product
In the windows “Primer Size” the minimal (“Min”) and maximal (“Max”) primer length values are defined, as well as the optimal (“Opt”) value.
6. According to the default definitions, what should be the length of the primers?
- Minimal primer length: 18 nucleotides, maximal: 20 nucleotides, optimal: 27 nucleotides.
- Minimal primer length: 18 nucleotides, maximal: 27 nucleotides, optimal: 20 nucleotides.
- Minimal primer length: 27 nucleotides, maximal: 20 nucleotides, optimal: 18 nucleotides.
- Minimal primer length: 57 nucleotides, maximal: 63 nucleotides, optimal: 60 nucleotides.
The answer is: B. The default definitions of the tool are primers of 18 to 27 nucleotides length, likely 20 nucleotides.
Primers of this length were found to be specific, meaning the probability that they will align to the complement DNA sequence only, and not other DNA segments, is high.
In the windows “Primer Tm” the minimal (“Min”), optimal (“Opt”), and maximal (“Max”) annealing temperatures are defined. The annealing temperature is the temperature in which half of the primers binds to their complementary sequences; it will be possible mainly at the annealing temperature or below it.
Teachers: The opposite process from the annealing of the primer to the template DNA strand is melting. Melting temperature is the temperature in which half of the primers will dissociate from their complementary sequences. In temperatures lower than the melting temperature, more primers are adhered to the template strands. Pay attention that the definition refers in general to primers’ complementary sequences, rather than specifically to template DNA strands. Similarly, melting temperature also refers to double-stranded DNA, such as DNA plasmid or even the PCR product, where the two strands dissociate from each other.
7. What are the parameters that set the annealing temperature of each primer?
- The location of the primer on the DNA template.
- The length and sequence of the primer, meaning, the percentage of G and C nucleotides.
- The length of the expected amplification product.
- The optimal temperature for the polymerase activity.
The answer is: B. Both the length and sequence of the primer set its annealing temperature. For a primer that consists of more G and C nucleotides the annealing temperature should be higher since G and C nucleotides form 3 hydrogen bonds with nucleotides of the complementary strand (while A and T nucleotides form only 2 hydrogen bonds with nucleotides of the complementary strand). Similarly, for longer primers, a higher annealing temperature should be defined, since more nucleotides form bonds with the template strand. Therefore, the primer sequence and its length affect the annealing temperature. The default tool definitions indicate a temperature range of 57°-63°C, preferably 60°C.
The minimal (“Min”), optimal (“Opt”) and maximal (“Max”) proportion of guanine (G) and cytosine (C) type nucleotide in the primer sequence can be set in the windows “Primer GC%”. For this parameter, the definitionsare more flexible and usually allow primers consisting of 20% to 80% G or C nucleotides
8. Why is it important to define the percentage of G and C nucleotides (GC%) in the primer?
- Because it dictates the strength of the interaction between the primer and the DNA template (G and C form 3 hydrogen bonds while A and T form only 2 hydrogen bonds).
- Because it determines the strength of the bond between the two primers (G and C form 3 hydrogen bonds while A and T form only 2 hydrogen bonds).
- Because it determines the strength of the bond between the primer and the RNA template (G and C form 3 hydrogen bonds while A and T form only 2 hydrogen bonds).
- Because G and C are more prevalent in nature and are thus cheaper to use.
The answer is: A. Because it dictates the strength of the interaction between the primer and the DNA template (G and C form 3 hydrogen bonds while A and T form only 2 hydrogen bonds).
9. For which of the following parameters should the two primers be similar and why?
- Both primers should be of the same length so they can anneal to each other completely.
- Both primers should have the same percentage of G and C nucleotides (GC%) so they can bind the same region in the DNA template.
- Both primers should have the same annealing temperature (Tm) so that each primer will anneal to the complementary template at the same temperature.
- It is important that the sequence of one primer complements the sequence of the second primer so that they could adhere together.
The answer is: C. It is important that both primers will have the same annealing temperature (Tm) so that each primer will bind the template at the same temperature.
Teachers: At this point, it is important to emphasize again that the primers bind the DNA template, each to its specific complementary site rather than to each other. If the primers bind each other, the desired target sequence will not be amplified. The conditions in which each primer anneals to the template should be similar, to ensure optimal annealing of both primers at the same time. The length of the primer and its sequence (the percentage of the G and C nucleotides and their order) define the annealing temperature for each primer. This temperature will determine the PCR annealing temperature. It should also be mentioned that the tool designs several pairs of primers, each answers the default settings. The researchers should choose a pair of primers that fits their needs.
In the “Product Size” window it is possible to define the length of the expected amplification product, meaning the length of the DNA segment that lies between the two primers. The default setting of the tool is to design primers aimed to amplify a product of 150-200 nucleotides. This length is long enough to ensure specific sequence identification on the one hand, and short enough to allow a fast and effective PCR reaction. The default definitions that set the characteristics of the primers and the expected amplification product can be modified, but since they are set to design optimal primers we are not obliged to change them.