We found the active site in the IPNS protein and discovered that all three amino acids comprising the active site are close to each other in the spatial structure. Now we wish to understand what is the substrate that binds to the active site, how is the active site adjusted to its shape and what is the product of the IPNS protein’s activity.
The IPNS enzyme catalyzes the process in which a long molecule called ACV converts into a molecule called IPN (Figure 11). The IPN molecule is part of the creation process of the antibiotics penicillin and cephalosporin, and therefore it is of high importance.
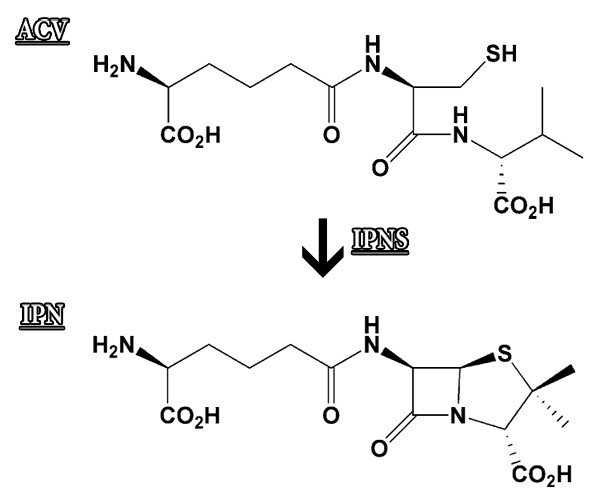
13. What obvious difference can you see between the ACV substrate molecule and the molecule of the product IPN?In order to decipher the spatial structure of the protein, we must first produce protein crystals. The crystallization solution of the IPNS protein that we are viewing, contained substrate molecules as well as product molecules; therefore, some of the protein crystals created contain the substrate molecule and some contain the product molecule, and we can see them in the spatial structure of the protein complex.Now we will return to the Jmol workspace and display the substrate molecule (ACV). This action requires two stages here as well: in the first stage we select the ACV substrate molecule with the Select command, and in the second stage we display the bonds between the atoms of the ACV molecule in a visible way (Figure 12).
Figure 12: The stages for displaying the bonds between the atoms of the ACV substrate molecule.
- In the workspace right click the mouse to open the additional options menu.
- Pick the command Select ➔ Hetero ➔ By Hetatm ➔ ACV – L-D-(A-AMINOADIPOYL)… (at this stage the model has not changed).
- Right click the mouse and pick the command Style ➔ Bonds -> 0.3Å
- Right click the mouse and pick the command Color ➔ Bonds -> Red
Now the substrate molecule is displayed in red, and the active site is displayed in cyan (screen 10).
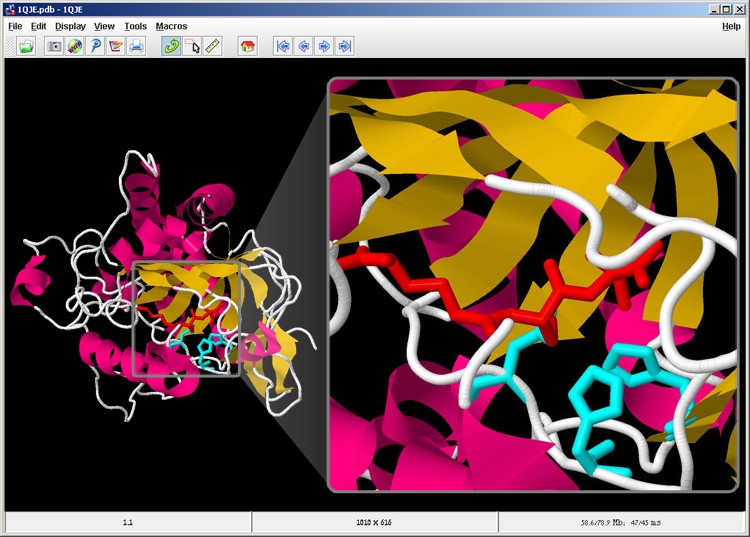
Screen 10: The substrate molecule (red) is located opposite the active site (cyan) in the IPNS enzyme
Using a similar method, we will display the product molecule in violet.
Right click the mouse in the workspace. Open the additional options menu.
- Pick Select ➔ Hetero ➔ By Hetatm ➔ IP1 isopenecillin N (at at this stage the model has not changed).
- Right click the mouse and pick the command Style ➔ Bonds ➔ 0.3Å
- Right click the mouse and pick the command Color ➔ Bonds ➔ Violet
- Right click the mouse and pick the command Zoom ➔ 200%
Now the product molecule is also displayed in violet (screen 11).
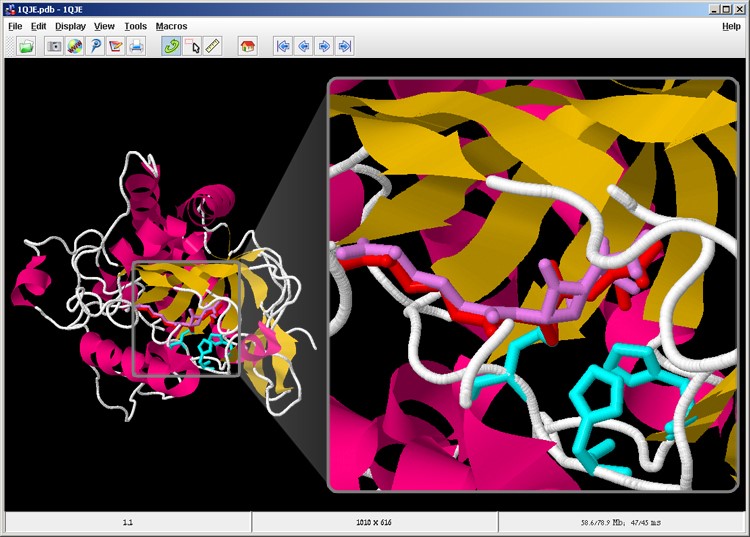
Screen 11: The substrate molecule (red) and the product molecule (violet) are located opposite the active site (cyan) in the IPNS enzyme.
14. Rotate the model and observe the substrate and the product. What is the main difference between these two molecules?
It should be emphasized again that after the IPNS enzyme binds to the ACV substrate, the IPN product molecule produced has a different structure than the substrate, therefore the IPN product detaches from the enzyme. That is, the enzyme is not attached to the substrate and the product simultaneously. When the crystals used to determine the spatial structure of the IPNS enzyme were created, some crystals were created with the enzyme attached to the product, and some were created with the enzyme attached to the product moments before detaching from it. This is why in the structure, the substrate as well as the product, and not just one of them, are visible in relation to the enzyme. An iron metal ion was also identified along with the protein structure.
15. Where do you think the iron metal ion is located in the IPNS protein? Explain your choice.
We will display the metal ion in the protein model. This action requires two stages as well. In the first stage we select the metal ion with the Select command, and in the second stage we change the volume of the metal ion so that it is visible in the model, in this way:
- In the workspace right click the mouse. Open the additional options menu
- Pick the command Select ➔ Hetero ➔ By Hetatm ➔ FE2- FE (II) ION (at this stage the model has not changed).
- Right click the mouse and pick the command Style ➔ Atoms ➔ 100% van der Waals
The metal atom is also displayed before us, and we can see that it is located between the three amino acids that comprise the active site on one side, and near the substrate molecule on the other side (screen 12). In this central location the iron ion serves as an electron receptor and accelerates the catalytic process.
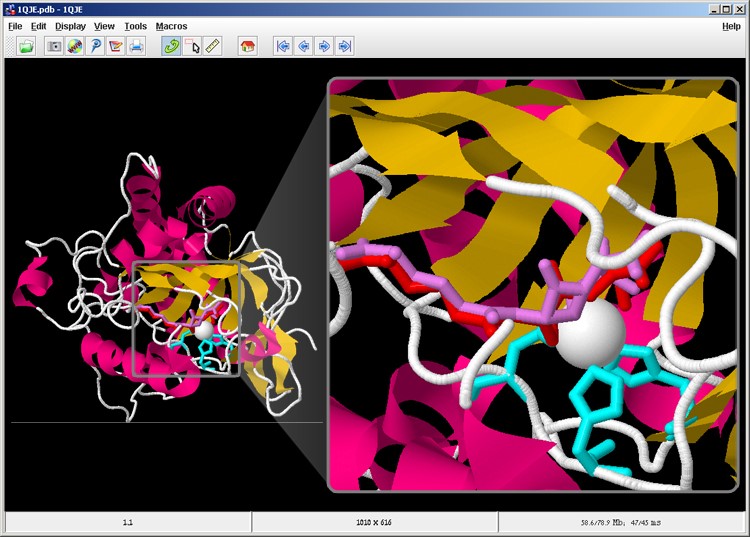
Screen 12: The metal ion (white) in relation to the active site (cyan) the substrate (red) and the product (purple) in the IPNS complex.
In this part we presented the three amino acids in the active site and the location of the substrate in relation to the active site. We saw that the metal ion serves as a cofactor, which accelerates the catalytic process, in which chemical bonds change in the substrate molecule so that a closed 5-membered ring is created. This reaction changes the structure of the molecule. The product molecule is shorter than the substrate and changes an angle in the area of the rings. Therefore, the product molecule detaches from the active site and is released to the watery surroundings.