A quick reminder…
To succeed in this assignment, you must be familiar with the following terms from basic genetics: DNA, gene, allele, genetic linkage , and from the genome and bioinformatics: polymorphic markers, genome mapping, genome sequencing.
Why is determining the gene's inheritance pattern not enough?
By the end of the genetic counseling, the only thing known about the gene is how it is inherited. If you know this, you can estimate which family members will be deaf in adulthood. But, as molecular geneticists, we want to know more. We know that in humans approximately 60 genes are involved in the hearing process. If we locate in the genome the particular gene involved in hearing loss in this family, we will be able to differentiate between the DNA sequences of hearing and deaf family members. This information can help in early diagnosis of family members who probably will be deaf in adulthood.
How does determining the gene's inheritance pattern help identify it in the genome?
In the genetic counseling assignment, you concluded that the deaf family members had one abnormal allele. Because this allele is passed on by inheritance in the family, it can be assumed that its DNA sequence is identical in all deaf members and different among the hearing adults. Therefore, the search in the genome is for an allele that is identical in all deaf members and different from that of the hearing members.
How is a gene identified in the genome?
Although you do not know where the gene of interest is located in the genome, a number of biological facts can help you identify it:
- In the family's genome there is one significant difference between the DNA sequences of the deaf and hearing adults: the allele whose defect causes deafness is identical in all deaf members and different in the hearing members.
- This allele is part of a "linkage group", that is, a group of alleles and nearby polymorphic markers, on the same DNA segment which are inherited along with it. They are linked to the gene of interest and will be identical in all deaf family members and different in the hearing family members.
- Polymorphic markers have been mapped in the Human Genome Project, meaning that the location of these markers in the genome is already known, and the DNA sequence of the different alleles of each polymorphic marker has been determined.
"Gene hunters" make use of these biological facts: instead of looking for the alleles of a gene of interest, they take an indirect approach. They look for polymorphic markers that are probably part of the linkage group of this allele of interest. Relying on the difference in the phenotype, a difference in the markers in the genotype can be found, thus narrowing the search for the gene to a new, smaller area.
The following diagram demonstrates the process of identifying a region in the genome containing a gene of interest, using polymorphic markers.
Look at the example below, which depicts a pedigree of a family in which the gene involved in hearing is inherited. The allele determining deafness is dominant and is marked with the letter D. The two parallel lines for each family member represent a pair of segments from homologous chromosomes, each of which contains one allele of the gene.
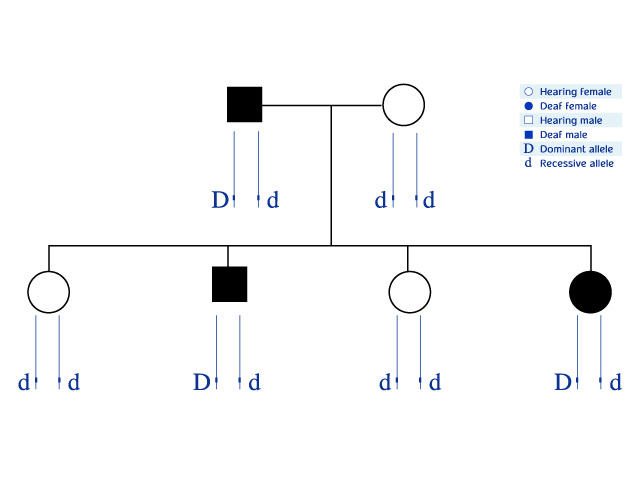
The DNA sequence of the polymorphic marker is on the same chromosome segment, linked to the gene involved in hearing (![]() ). In this family, there are three different alleles of the marker (1,2,3
). In this family, there are three different alleles of the marker (1,2,3 ![]() ).
).
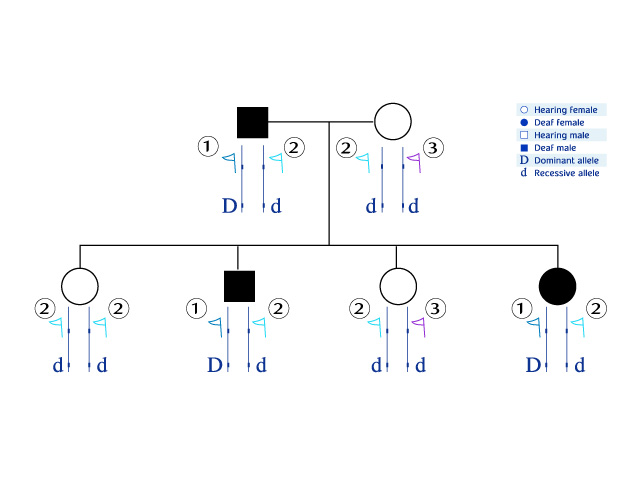 Note that in this family the D allele (for deafness) is strongly linked to allele 1
Note that in this family the D allele (for deafness) is strongly linked to allele 1![]() In other words, this family has a D allele and 1
In other words, this family has a D allele and 1 ![]() allele which are inherited together (allele d can be linked to alleles 2
allele which are inherited together (allele d can be linked to alleles 2 ![]() or 3
or 3 ![]() ).
).
On the same chromosome segment, further from the gene of interest, there is another polymorphic marker (![]() ). In this family there are four different alleles of this marker (3,5,6,7
). In this family there are four different alleles of this marker (3,5,6,7 ![]() ). This marker is weakly linked to the gene involved in hearing and to the marker
). This marker is weakly linked to the gene involved in hearing and to the marker ![]() .
.
 Note that the chromosome segment containing the D allele of the deaf daughter is different from the segment containing the D allele of the deaf father (marker 5
Note that the chromosome segment containing the D allele of the deaf daughter is different from the segment containing the D allele of the deaf father (marker 5 ![]() in the daughter instead of marker 3
in the daughter instead of marker 3 ![]() in the father). The source of the difference, most probably, is an example of crossing-over event between the father's chromosomes during gametogenesis. In this process, the markers 3
in the father). The source of the difference, most probably, is an example of crossing-over event between the father's chromosomes during gametogenesis. In this process, the markers 3 ![]() and 5
and 5 ![]() were swapped in the father.
were swapped in the father.
In our demonstration, the location of the D gene and the markers linked to it are known. In reality, however, the researcher has no idea where the gene is in the genome, and certainly does not know what chromosome it is on. But he can identify a gene based on signs of the polymorphic markers linked to it:
Imagine a researcher who receives blood samples of all the family members from a medical laboratory. In his molecular research laboratory, he isolates DNA from each sample, and using biochemical methods he checks to see which of the alleles of known polymorphic markers can be found in the genome of each individual. There are actually many polymorphic markers in the human genome. For the sake of simplicity, we have chosen to display the information obtained for only two markers:
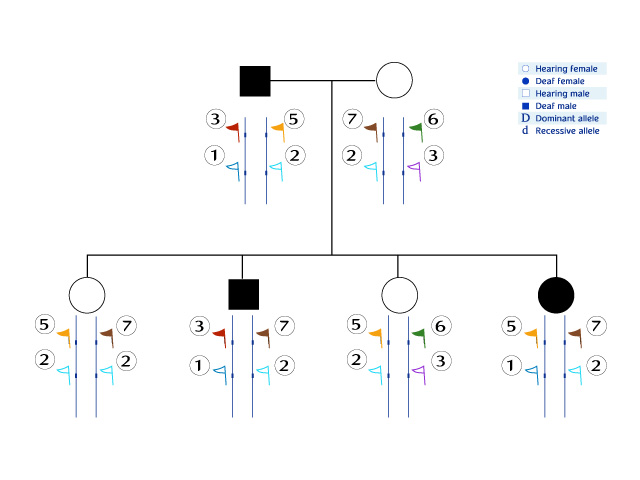 A researcher who has obtained this picture may be very close to identifying a gene of interest:
A researcher who has obtained this picture may be very close to identifying a gene of interest:
- He knows the phenotype of each of the family members.
- He has checked the alleles of the polymorphic markers of each of the family members.
- The only thing left for him to do is to look for alleles of markers that differentiate the deaf from the hearing individuals, i.e. the group of markers that have alleles of the same type in all the deaf family members exclusively.
- Which polymorphic marker allele can be found only in the deaf individuals and not in the hearing ones in this family?
- What is the significance of such an allele for the researcher? Note that the crossing-over process reduces the size of the search region: the father and daughter have different markers for this allele and they are both deaf. This means that the gene of interest is probably not next to the marker that has crossed over.
And now for the true story…
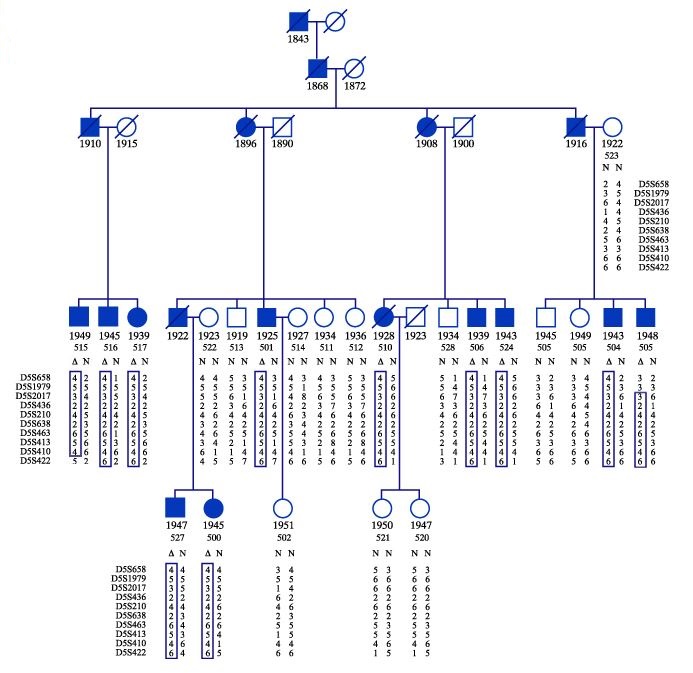
Let's look at the pedigree that you saw in the previous assignment. A molecular biologist screened the genome of each of the family members to identify the various polymorphic marker alleles. He found (using statistics) a DNA segment on chromosome 5 whose polymorphic markers are identical among all the deaf adults and different among the hearing adults in the family. Markers from this segment are indicated in the pedigree.
The names appearing alongside the chart are the names of the polymorphic markers that are located near each other on the specific segment of human chromosome 5. For example: the top marker is called D5S658, next to it is D5S1979, followed by D5S2017, according to the DNA sequence.
The numbers appearing in the two columns under each individual in the family indicate the polymorphic marker allele pairs. A pair of parallel numbers along each segment indicates the pair of marker alleles whose name appears on the left of the diagram. For example: the D5S658 marker in individual number 517 has alleles 2 and 4, whereas the same marker in individual 516 has alleles 1 and 4. The numbers indicate different sequences, characteristic of the normal polymorphism in the population (a single base difference is enough).
In Assignment 1 you found that the deafness allele was dominant. It is enough for an individual to have one such allele of the gene involved in hearing in order to be deaf.
- In the simple pedigree presented earlier, you identified an allele of a polymorphic marker that appears only in deaf and not in hearing individuals. This allele was linked to the gene involved in hearing.
In the pedigree below, track the markers on the deaf individuals' chromosome segment. Find the chromosome segment whose combination of polymorphic marker alleles appears in all the deaf individuals. Compare this segment with the chromosome segments of the hearing population. The combination you found in the deaf individuals should not appear in any of the hearing individuals. This marker group is probably linked to the gene involved in hearing.
Note: In all of the deaf individuals, one of the chromosome segments should be identical, whereas its homolog should be different. In the hearing individuals, the segments in the two homologous chromosomes are, of course, different. - Look at the markers which appear in all the deaf individuals:
- Are the markers identical in all the deaf individuals?
- The size of the common segment differs among the deaf individuals. It varies due to crossing-over events between alleles. A "gene hunter" stated that crossing-over is sometimes helpful to identify a gene in the genome. Try to explain what this scientist meant.
- State the names of the markers which border the smallest DNA segment that probably contain the gene of interest.
What have you learned from this assignment?
You have learned that once the most likely inheritance pattern of a family disorder has been determined, an attempt can be made to identify the gene in the genome for the purpose of diagnosis or curing. For this purpose, molecular geneticists compare the genome of the family members using a DNA segment that is identical in all the affected individuals and different in all those with the normal phenotype.
As molecular geneticists, you have made use of a genetic linkage marker map to identify a region of the genome containing identical alleles of genetic markers in all the deaf family members, and that are different in the hearing adult members.
In the particular case that you have examined, you identified a group of genetic polymorphic markers that are linked to the gene involved in hearing. Identification was made possible by tracking the family phenotype and locating a whole segment of a chromosome, containing a combination of a number of alleles of different polymorphic markers, that exists only in the deaf individuals and not in the hearing ones. This marker group is linked to the gene involved in hearing.
We can therefore assume that the gene involved in hearing, whether normal or abnormal, is hidden somewhere in the DNA segment containing these markers.

